Ex. 5.10
Ex. 5.10
Derive an expression for \(\text{Var}(\hat f_\lambda(x_0))\) and \(\text{Bias}(\hat f_\lambda(x_0))\). Using the example (5.22), create a version of Figure 5.9 where the mean and several (pointwise) quantiles of \(\hat f_\lambda(x)\) are shown.
Soln. 5.10
Since
\[\begin{equation}
\hat{\mathbf{f}} = \bb{N}(\bb{N}^T\bb{N} + \lambda \bm{\Omega}_N)^{-1}\bb{N}^T\by\non
\end{equation}\]
we have
\[\begin{equation}
\hat f_\lambda(x_0) = \bb{N}(x_0)^T(\bb{N}^T\bb{N} + \lambda \bm{\Omega}_N)^{-1}\bb{N}^T\by\non
\end{equation}\]
where \(\bb{N}(x_0) = (N_1(x_0), ..., N_N(x_0))^T\). Therefore we have
\[\begin{eqnarray}
\text{Var}(\hat f_\lambda(x_0)) &=& \bb{N}(x_0)^T(\bb{N}^T\bb{N} + \lambda \bm{\Omega}_N)^{-1}\bb{N}^T\text{Var}(\by)(\bb{N}^T\bb{N} + \lambda \bm{\Omega}_N)^{-1}\bb{N}(x_0)\non
\end{eqnarray}\]
and
\[\begin{eqnarray}
\text{Bias}(\hat f_\lambda(x_0)) &=& f_\lambda(x_0) - E[\hat f_\lambda(x_0)]\non\\
&=&f_\lambda(x_0) - \bb{N}(x_0)(\bb{N}^T\bb{N} + \lambda \bm{\Omega}_N)^{-1}\bb{N}^T\by.\non
\end{eqnarray}\]
Next we reproduce Figure 5.9 in the text. Since smoothing splines has an explicit, finite-dimensional, unique minimizer which is a natural cubic spline with knots at the unique values of the \(x_i\), we could use codes for natural cubic splines (I haven't found a convenient package for smoothing splines in Python yet.)
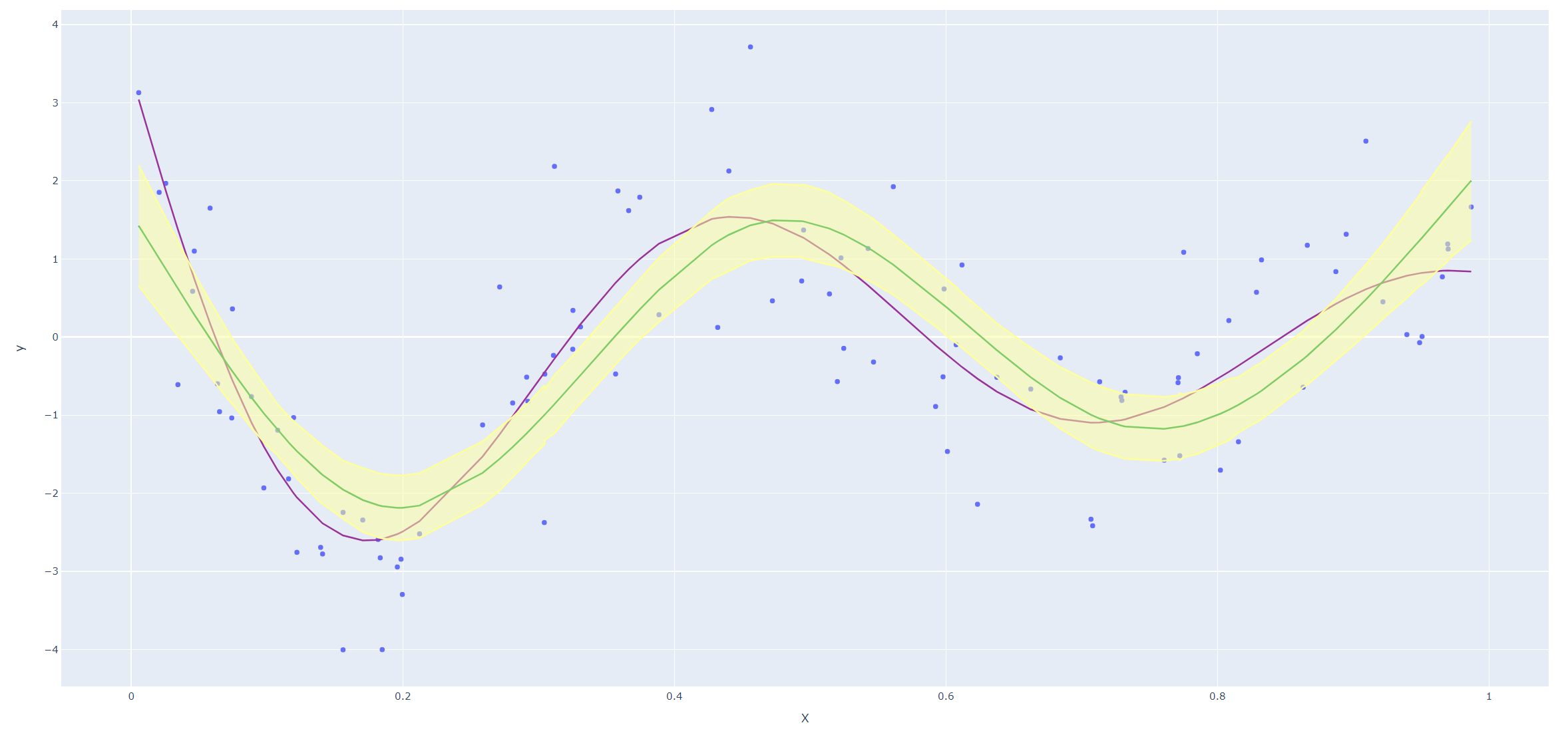
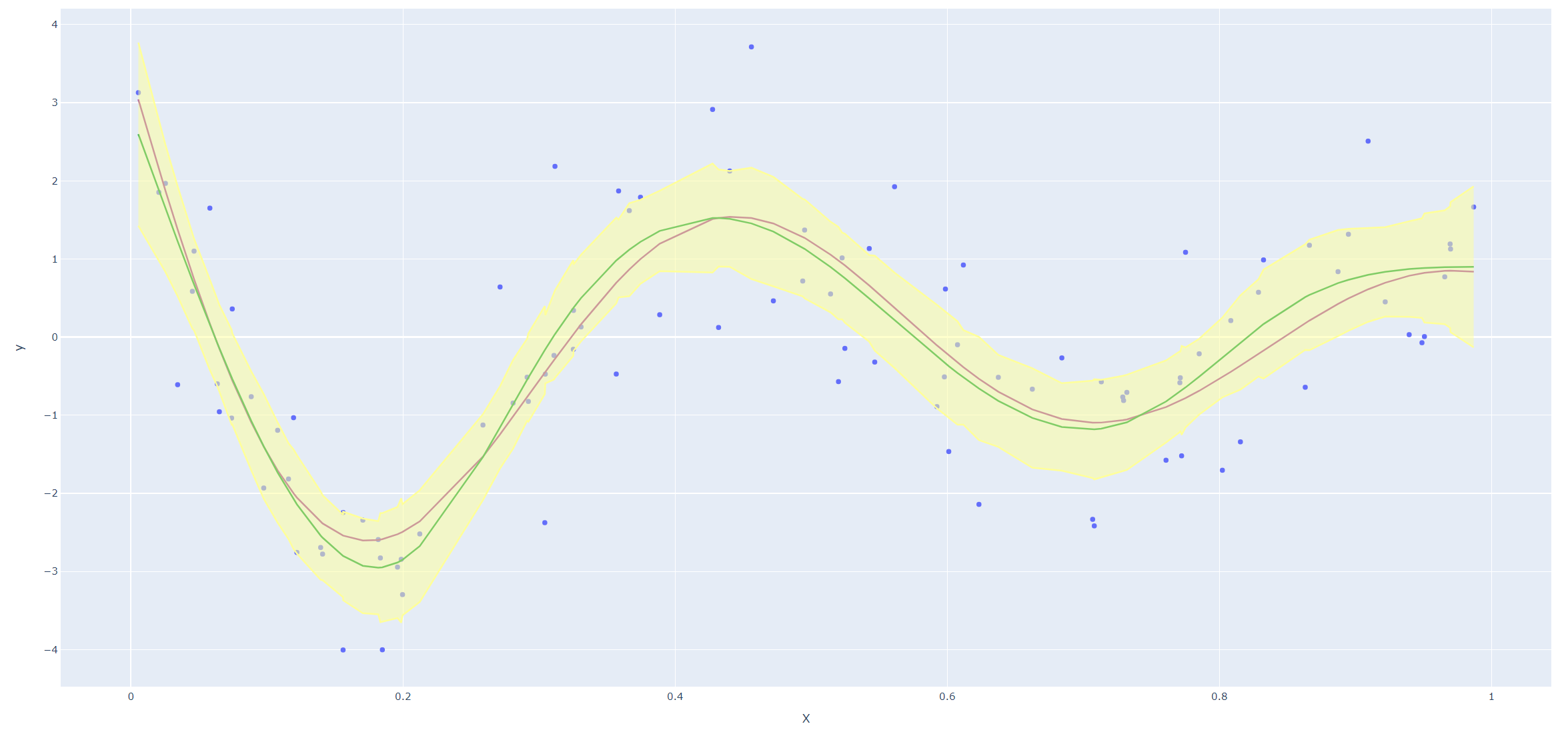
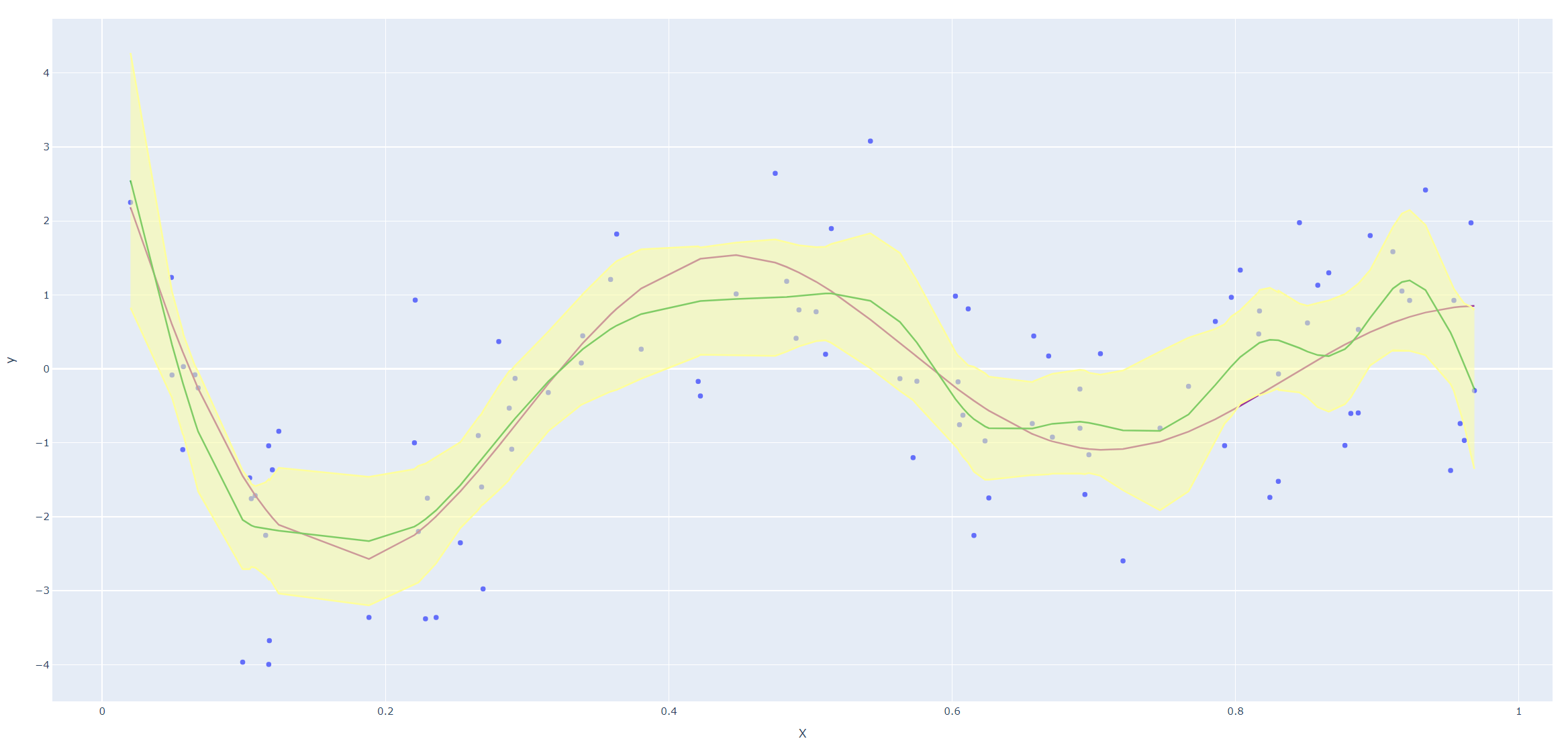
Code
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 | |