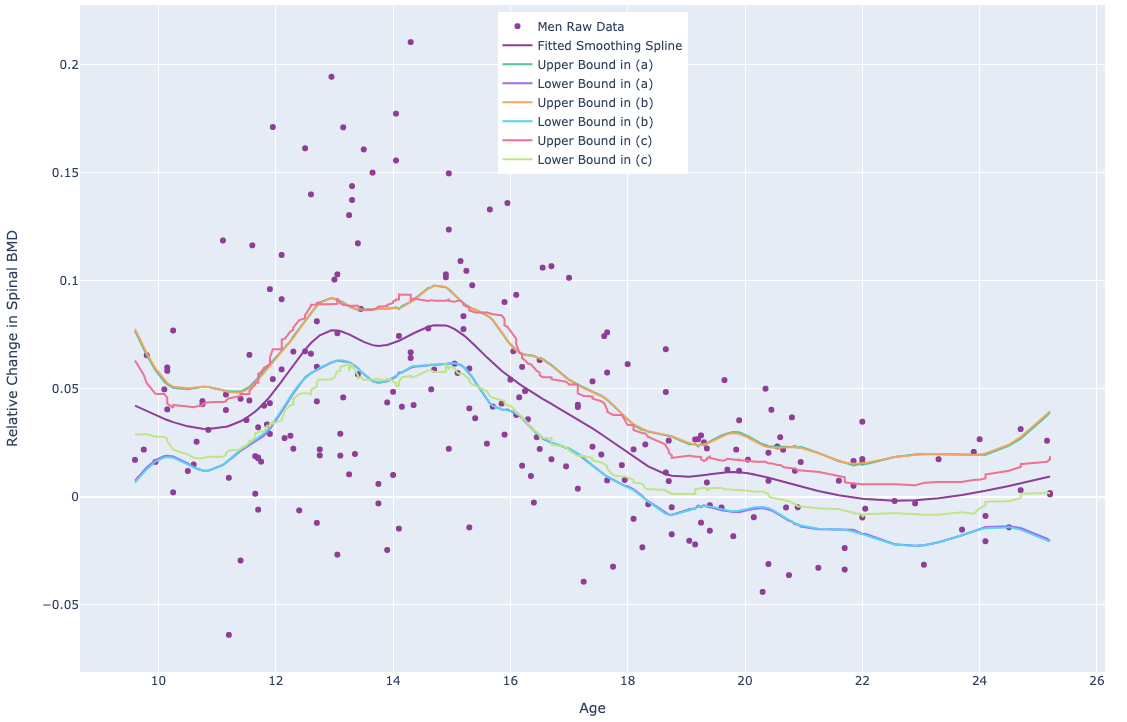
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146 | import pathlib
import numpy as np
import pandas as pd
from numpy.linalg import inv
from numpy.linalg import multi_dot
from sklearn.base import BaseEstimator
from sklearn.model_selection import GridSearchCV
from sklearn.metrics import mean_squared_error
from sklearn.utils import resample
import statsmodels.api as sm
from patsy import dmatrix
import plotly.graph_objects as go
# get relative data folder
PATH = pathlib.Path(__file__).resolve().parents[1]
DATA_PATH = PATH.joinpath("data").resolve()
# prepare data
data = pd.read_csv(DATA_PATH.joinpath("boneMineralDensity.csv"), header=0)
data_men = data.loc[data['gender'] == 'male']
data_men = data_men.sort_values('age')
X_men = data_men.loc[:, 'age'].to_numpy()
y_men = data_men.loc[:, 'spnbmd'].to_numpy()
data_women = data.loc[data['gender'] == 'female']
data_women = data_women.sort_values('age')
X_women = data_women.loc[:, 'age'].to_numpy()
y_women = data_women.loc[:, 'spnbmd'].to_numpy()
# define a cubic smooth spline estimator
class CubicSmoothSpline(BaseEstimator):
def __init__(self, df=10):
self.df = df
self.H = None
self.fitNatural = None
self.pred = None
def fit(self, X, y=None):
self.H = dmatrix('cr(x, df={})'.format(self.df), {'x': X}, return_type="dataframe")
self.fitNatural = sm.GLM(y, self.H).fit()
return self
def predict(self, X):
self.pred = self.fitNatural.predict(dmatrix('cr(xp, df={})'.format(self.df), {'xp': X}))
return self.pred
# (a) cross validation
param_grid = [{'df': np.arange(5, 21)}]
css = CubicSmoothSpline()
grid_search = GridSearchCV(css, param_grid, cv=10, scoring='neg_mean_squared_error')
grid_search.fit(X_men, y_men)
final_model = grid_search.best_estimator_
final_df = final_model.df
print("The degree of freedom chosen by 10-fold CV is: {}".format(final_df))
# calculate point-wise variance
final_model.fit(X_men, y_men)
final_model.predict(X_men)
y_men_pred = final_model.pred
sigma_square = mean_squared_error(y_men_pred, y_men)
H = np.asarray(final_model.H)
m_Sigma = sigma_square * (inv(np.matmul(H.transpose(), H)))
m_nc = multi_dot([H, m_Sigma, H.transpose()])
pt_var_nc = m_nc.diagonal()
pt_std_nc = np.sqrt(pt_var_nc)
upper = y_men_pred + 1.65 * pt_std_nc
lower = y_men_pred - 1.65 * pt_std_nc
# plot
fig = go.Figure()
fig.add_trace(go.Scatter(x=X_men, y=y_men,
mode='markers',
name='Men Raw Data',
line_color='#993399'))
fig.add_trace(go.Scatter(x=X_men, y=y_men_pred,
mode='lines',
name='Fitted Smoothing Spline',
line_color='#993399'))
fig.add_trace(go.Scatter(x=X_men, y=upper,
mode='lines',
name='Upper Bound in (a)'))
fig.add_trace(go.Scatter(x=X_men, y=lower,
mode='lines',
name='Lower Bound in (a)'))
# (b): from (8.28) with tau=10 and \Sigma = Identity matrix
tau = 10
n = H.shape[1]
m_Sigma_2 = sigma_square * (inv(np.matmul(H.transpose(), H) + sigma_square / tau * np.identity(n)))
m_nc_2 = multi_dot([H, m_Sigma_2, H.transpose()])
pt_var_nc_2 = m_nc_2.diagonal()
pt_std_nc_2 = np.sqrt(pt_var_nc_2)
upper_2 = y_men_pred + 1.65 * pt_std_nc_2
lower_2 = y_men_pred - 1.65 * pt_std_nc_2
fig.add_trace(go.Scatter(x=X_men, y=upper_2,
mode='lines',
name='Upper Bound in (b)'))
fig.add_trace(go.Scatter(x=X_men, y=lower_2,
mode='lines',
name='Lower Bound in (b)'))
# (c) bootstrap method
B = 100
fitted_curve_list = []
for b in np.arange(B):
data_sample = resample(data_men, replace=True)
data_sample = data_sample.sort_values('age')
X_sample = data_sample.loc[:, 'age'].to_numpy()
y_sample = data_sample.loc[:, 'spnbmd'].to_numpy()
final_model.fit(X_sample, y_sample)
y_pred = final_model.predict(X_sample)
fitted_curve_list.append(y_pred)
bs = np.stack(fitted_curve_list)
upper_3 = np.percentile(bs, 90, axis=0, interpolation='nearest')
lower_3 = np.percentile(bs, 10, axis=0, interpolation='nearest')
fig.add_trace(go.Scatter(x=X_men, y=upper_3,
mode='lines',
name='Upper Bound in (c)'))
fig.add_trace(go.Scatter(x=X_men, y=lower_3,
mode='lines',
name='Lower Bound in (c)'))
fig.update_layout(
xaxis_title="Age",
yaxis_title="Relative Change in Spinal BMD",
)
fig.update_layout(legend=dict(
yanchor="top",
y=0.99,
xanchor="center",
x=0.5
))
fig.show()
|